This work was done in collaboration with Mahendra Khened and Vikas Kumar Anand
Introduction
Medical imaging, specifically radiologic imaging is the most commonly used diagnostic tool for disease diagnosis and treatment assessment for a wide variety of conditions. Over the last decades these imaging systems with improved hardware for image acquisition and sophisticated techniques for image reconstruction or estimation, provides increasingly complex data for radioilogists to sif through and provide an accurate diagnosis and guidelines. The complexity here is not only limited to In this article, we will be looking at what is biomedical imaging, the applications of radiology and histopathology as use-cases of biomedical imaging, and how deep learning is aiding the analysis of biomedical images towards early and more accurate diagnosis.
What is Biomedical Imaging?
Biomedical Imaging has several disciplines such as medical Imaging, molecular imaging, fluorescent imaging, optical imaging, and microscopy (Digital Histopathology). This article is mainly focused on medical imaging and digital histopathology. Medical imaging consists of a series of processes or techniques for the clinical monitoring, diagnosis, and treatment of diseases and injuries to establish visual representations of the internal sections of the body, such as organs or tissues. Moreover, it also helps to build anatomy and physiology databases. Thanks to today’s developments in the area, medical imaging has the capacity to collect human body knowledge for many useful clinical applications. Various forms of medical imaging equipment offer various details about the location of the body to be examined or medically treated. Medical imaging has different modalities such as Radiography, Tomography, Ultrasound, Magnetic Resonance Imaging (MRI), Nuclear medicine imaging (like PET,SPECT), Colonoscopy, and Mammogram imaging.
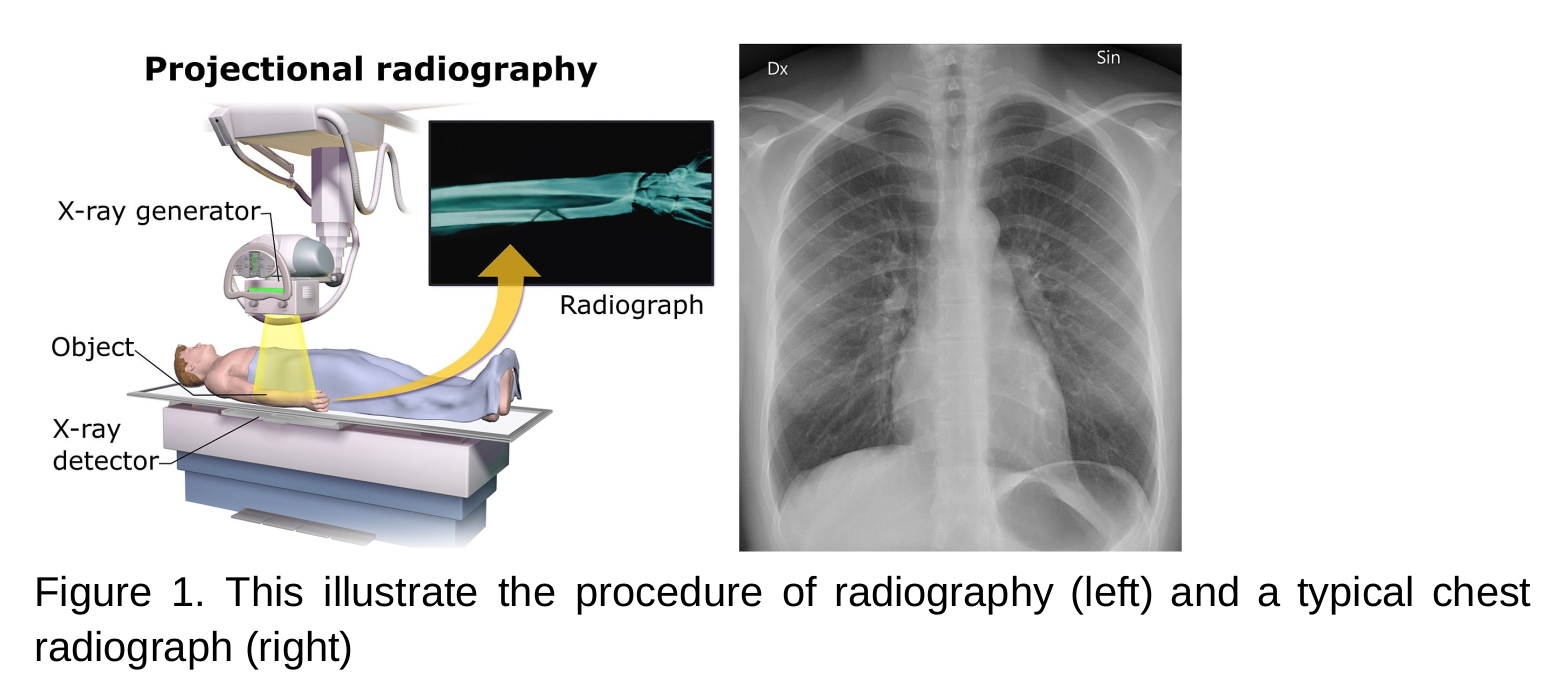
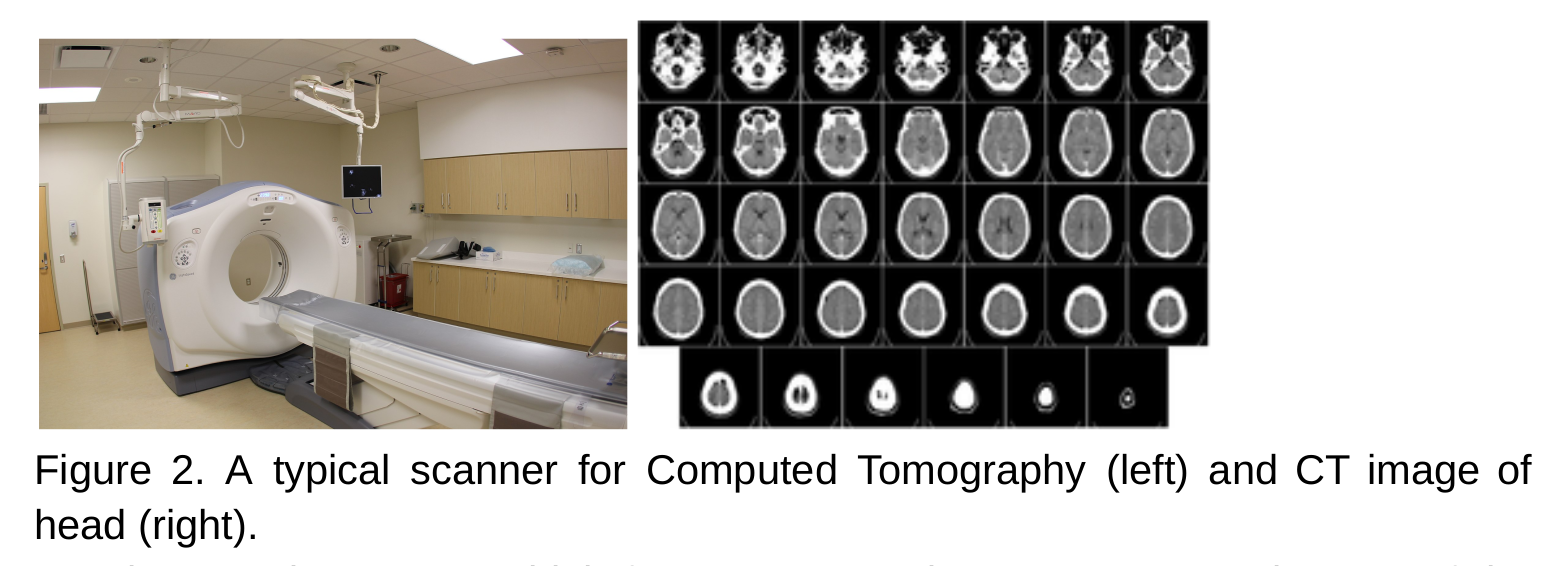
- Radiography and tomography uses x-ray to visualise the internal organs, tissues, and bones.
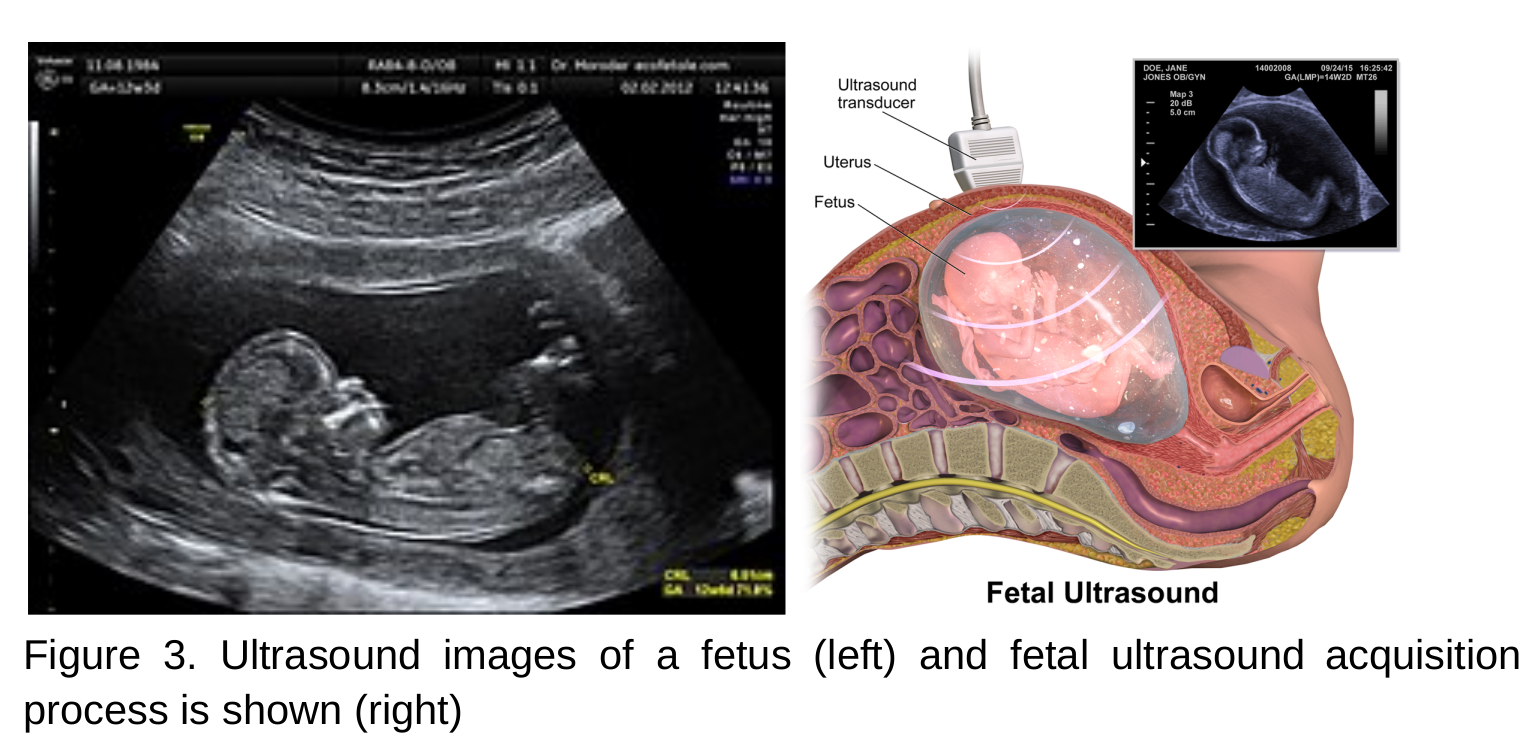
- An ultrasound scan uses high frequency sound waves to create images of the inside body.
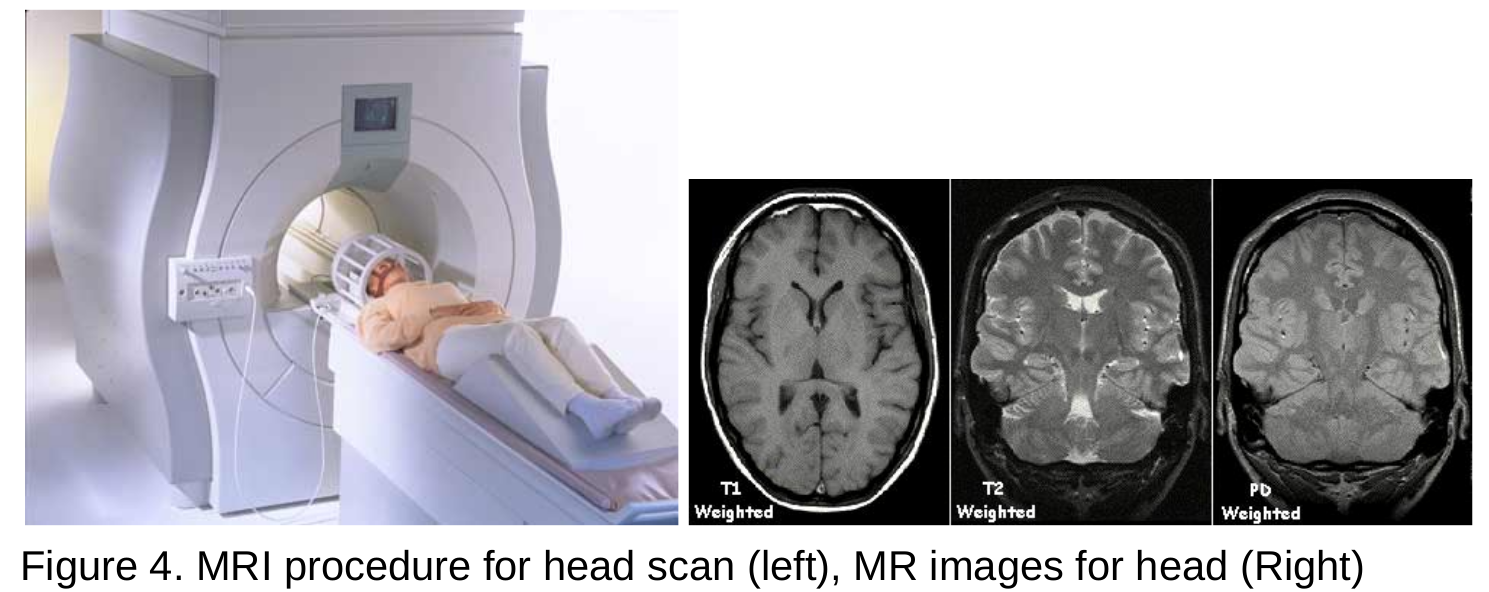
- MRI scanners use strong magnetic fields, magnetic field gradients, and radio waves to generate images of internal organs, and tissues.
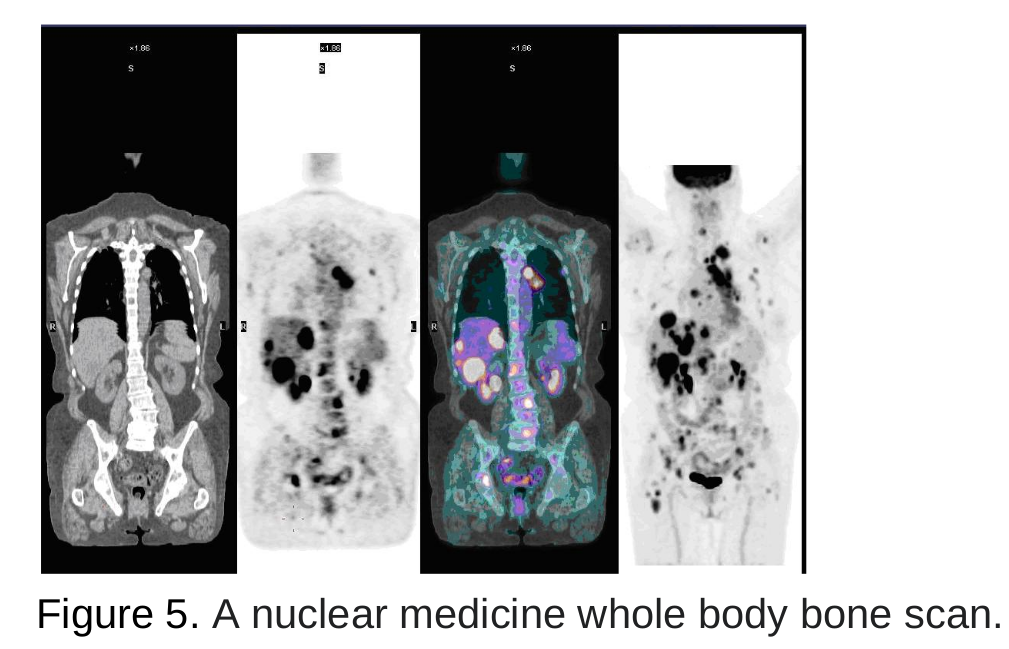
- Nuclear medicine imaging creates images by recording the radiation emitting from within the body. Nuclear medicine images are functional images.
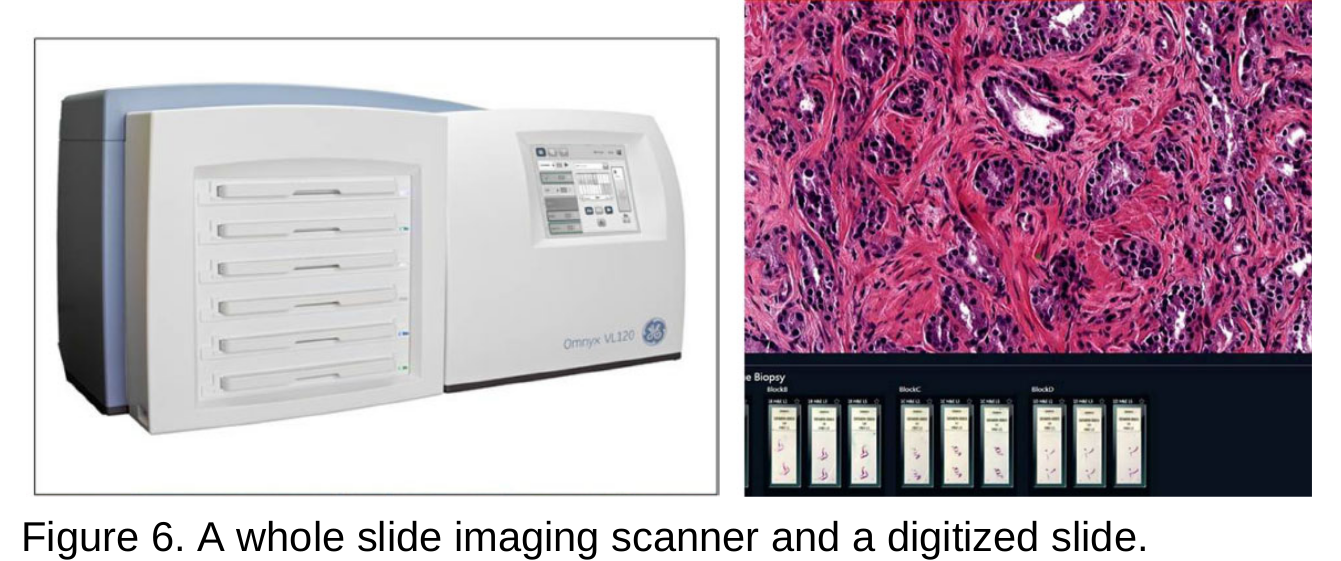
- Pathology is a medical discipline that deals with the study for disease diagnosis of tissue, cell and body fluid samples. Histopathology refers, in particular, to the microscopic examination of a tissue biopsy or surgical specimen that has been treated histologically and placed on glass slides. Digital pathology refers to the method of digitising glass slides using specialised scanners, then preserving them, processing and exchanging them with others. Digital pathology uses the technology of virtual microscopy or whole slide imaging (WSI), which refers to the method of scanning glass histology slides for examination on a computer screen at a sufficiently high resolution.
Importance of Medical Imaging
The use of medical imaging for patient care is considered to be a vital validation of many diseases and illnesses being examined and reported. High-quality imaging improves decision-making and may eliminate medical procedures that are unnecessary. Early treatment involving exploratory tests to assess the elderly and children can be avoided. Vital health records can be readily made accessible from time to time with the advancement of medical imaging, which can help detect diseases such as pneumonia, cancer, internal bleeding, brain tumor, and much more. Morphological descriptions are recorded in radiology and pathology on various biological scales. In the field of oncology, radiology helps to detect suspected lesions and clinical stages and, on the other hand, pathology, which is known to be the gold standard for tumour evaluation and grading, characterises complex tissue histological and molecular characteristics. Computer-aided analysis and interpretation of medical image data are important in order to optimise the benefits of radiological and pathological studies.
Medical Image Analysis
Typical image processing operations include segmenting regions, detecting objects and classifying images. In downstream analyses that incorporate information from clinical and molecular data and establish predictive and correlation models, more image features derived from segmentation may be used.
- Radiomics is a methodology that uses algorithms for data characterization to extract large amounts of features from images of radiology. Radiomic characteristics may show disease characteristics which are not detected by the naked eye. Numerous medical image analysis research studies have led to the creation of innovative methodologies to turn raw medical image data into such rich knowledge and new understanding effectively, accurately and reliably.
- Much of the recent medical image analysis work has centred on the development of algorithms for machine learning and, in particular, deep learning models. For medical image analysis concerns, these models have tremendous potential to be gradually realised. In medical image processing, the usefulness of deep learning is doubled, namely in automated analysis and the discovery of information. Medical image segmentation and classification include some of the automated analysis processes in routine clinical diagnostics. The purpose of the information discovery is to identify patterns in the data, such as trends in data that may inform about diagnosis , prognosis, response to treatment and genomic characteristics. In the face of a rising array of research and development methods and techniques, computer-based image analysis is still a challenge. The complexity of both image resolutions and data continues to increase, including improving existing techniques and creating new techniques.
Machine Learning and Deep Learning
Machine learning is a sub-field of computer science that developed from the study of artificial intelligence and pattern recognition in computational learning theory. Machine learning explores the design of algorithms that learn from data and make data predictions. In order to construct a model that can later be used for making data-driven predictions or decisions, such algorithms apply statistical techniques to the training set of observations. Machine learning usually gives the computer the ability to learn from examples without being specifically programmed for a particular task to be performed. Deep Learning consists of a series of algorithms for machine learning, also known as deep neural networks ( DNNs), which over the past decade have achieved remarkable success in processing natural types of data, such as images , text, and voice. Deep learning attempts to solve tasks with the use of a hierarchic representation framework to learn abstraction of information using nonlinear transformation architectures. Before the deep learning revolution, for better results, most well-developed machine learning algorithms needed the creation of structured data forms. This needed, however, in order to identify patterns and trends in the data, deep domain knowledge in engineering features from the data for the learning algorithm. Deep learning, on the other hand, is known for its inherent ability to jointly learn the representation of associated features from the data for performing a task.
Application of Machine Learning and Deep learning in Medical image Analysis
This is an article about biomedical image analysis. Here, we have illustrated the use of machine learning and deep learning techniques in biomedical image analysis. First illustration deals with the analysis of medical images obtained from MRI. It explains the possibility of exploitation of deep learning algorithms for segmentation of different components of the heart (such as Left ventricle, Left Atrium, Right Ventricle, Right Atrium, and Myocardial wall) and usage of machine learning for prognosis of different cardiac conditions. Application of deep learning for analysis of whole slide images is taken as the second use case. Whole slide images are taken at different magnification levels that make these images very large in size. Processing such a large image itself is a challenge and application of deep learning algorithms on these images pose different challenges. How has these challenges been overcome, is explained in the second use case.
Use Case 1: Deep learning based framework for Cardiac Segmentation
Cardiac cine Magnetic Resonance Imaging (MRI) is primarily used for the assessment of cardiac function and diagnosis of Cardiovascular diseases (CVDs). Cardiac MRI is considered the most accurate method for the estimation of clinical parameters such as ejection fraction, ventricular volumes, stroke volume and myocardial mass. Delineating organs and structures from volumetric medical images, such as MR and computed tomography (CT) images, is usually considered the primary step for estimating clinical parameters, disease diagnosis, prediction of prognosis and surgical planning. We developed a deep learning based framework for cardiac segmentation which incorporated cardiac structures segmentation and cardiac disease diagnosis. The figure below illustrates the pipeline. The pipeline involved:
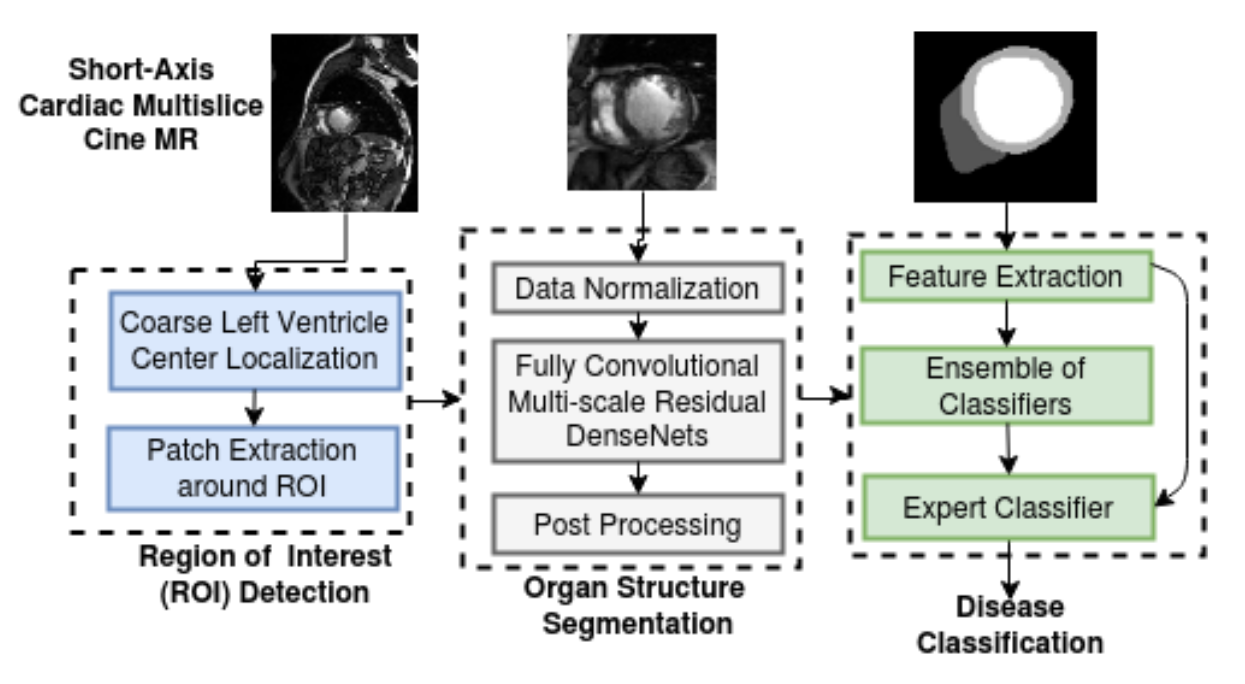
- Fourier analysis and circular Hough-transform for the region of interest (ROI) cropping: The cardiac MR images of the patient comprise the heart and the surrounding chest cavity like the lungs and diaphragm. The ROI detection step was explicitly designed to get an approximate localization of the heart region (LV centre) in cine MR images. ROI detection involved spatio-temporal statistical analysis of cardiac phases and circle Hough transform to delineate the heart structures from the surrounding tissues. The ROI extraction involved finding an approximate LV centre and extracting a patch of size 128x128 centred around it. The extracted ROI patch was used for training and inference of a fully convolutional neural network (FCN). This approach alleviates the class-imbalance problem associated with labels for heart structures seen in the full-sized cardiac MR images and also significantly reduces the GPU memory and model training time.
- FCN for cardiac structures segmentation: A typical semantic segmentation architecture comprises a down-sampling path (contracting) and an up-sampling path (expanding).
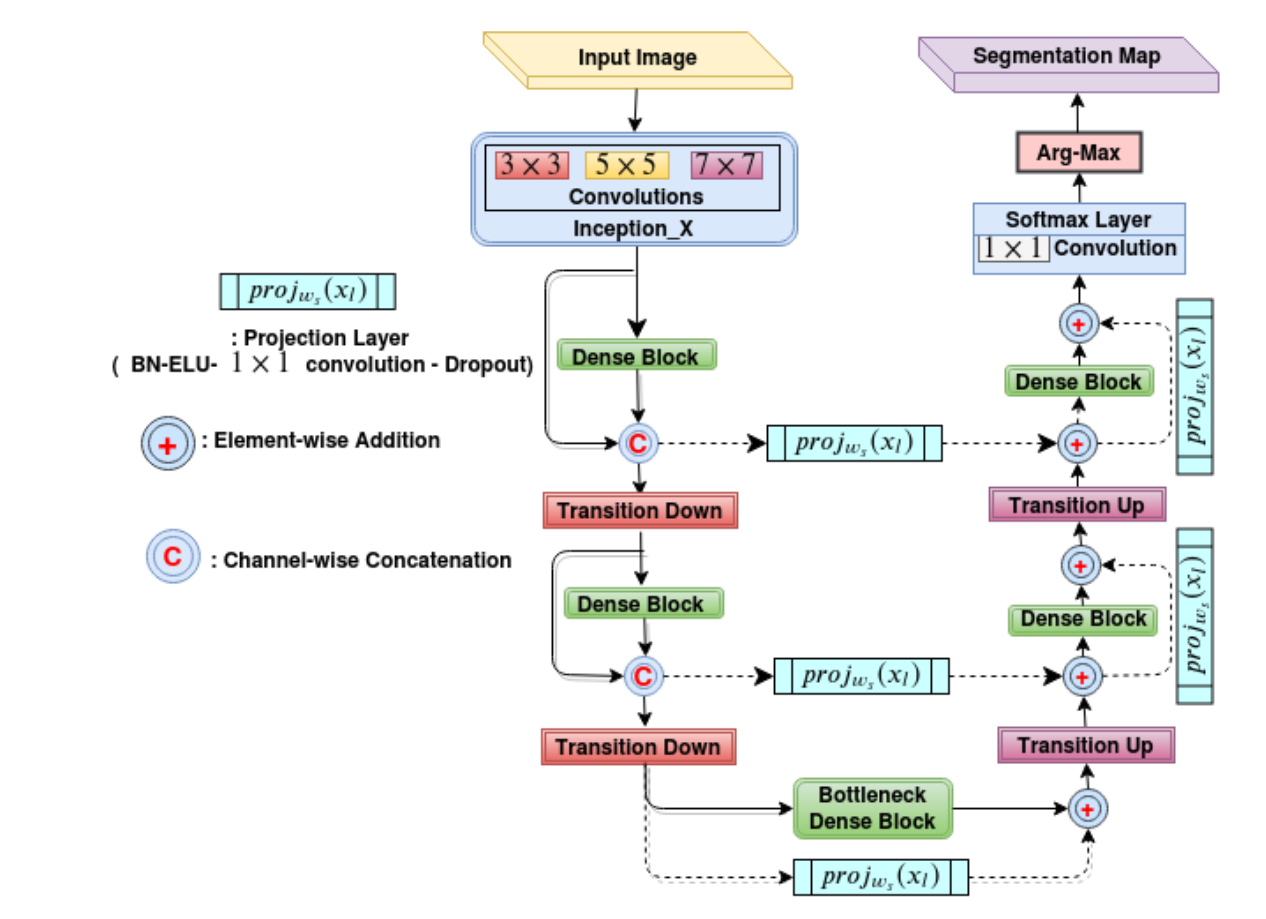 The figure above illustrates the schematic diagram of our FCN architecture. Our FCN’s connectivity pattern was based on DenseNets. Multi-scale processing was incorporated in the initial layers of the network by performing convolutions on the input with different kernel sizes in parallel paths and later fusing them as in Inception architectures. The down-sampling path of the network was similar to the DenseNet architecture. The last layer of the down-sampling path was referred to as the bottleneck layer. The input spatial resolution was recovered in the up-sampling path by transposed convolutions, dense blocks and skip connections coming from the down-sampling path. The up-sampling operation is referred to as transition up. The up-sampled feature maps were added element-wise with skip-connections. The feature maps of the hindmost up-sampling component were convolved with a 1x1 convolution layer followed by a soft-max layer to generate the final label map of the segmentation. The long skip and short-cut (residual) connections in the up-sampling path were computationally and memory-efficient when compared to standard skip connections based on copy and concatenation (as in U-Net). Our FCN design ensured significant reduction in the number parameters (by a factor of 100 times compared to the existing state-of-the-art architecture like U-Net for biomedical segmentation), and it was found to be suitable where there was a constraint on the availability of large annotated training datasets and computational resources. The FCN was trained using a custom loss function based on a weighted combination of cross-entropy and Dice loss.
The figure above illustrates the schematic diagram of our FCN architecture. Our FCN’s connectivity pattern was based on DenseNets. Multi-scale processing was incorporated in the initial layers of the network by performing convolutions on the input with different kernel sizes in parallel paths and later fusing them as in Inception architectures. The down-sampling path of the network was similar to the DenseNet architecture. The last layer of the down-sampling path was referred to as the bottleneck layer. The input spatial resolution was recovered in the up-sampling path by transposed convolutions, dense blocks and skip connections coming from the down-sampling path. The up-sampling operation is referred to as transition up. The up-sampled feature maps were added element-wise with skip-connections. The feature maps of the hindmost up-sampling component were convolved with a 1x1 convolution layer followed by a soft-max layer to generate the final label map of the segmentation. The long skip and short-cut (residual) connections in the up-sampling path were computationally and memory-efficient when compared to standard skip connections based on copy and concatenation (as in U-Net). Our FCN design ensured significant reduction in the number parameters (by a factor of 100 times compared to the existing state-of-the-art architecture like U-Net for biomedical segmentation), and it was found to be suitable where there was a constraint on the availability of large annotated training datasets and computational resources. The FCN was trained using a custom loss function based on a weighted combination of cross-entropy and Dice loss. - Automated cardiac disease classification: We developed an ensemble of classifiers trained on the features extracted from the segmentation map for developing automated diagnosis models. Based on cardiac physiological parameters in the medical reports, the patients are grouped into five classes namely (i) normal- NOR, (ii) patients with previous myocardial infarction- MINF, (iii) patients with dilated cardiomyopathy- DCM, (iv) patients with hypertrophic cardiomyopathy- HCM, and (v) patients with abnormal right ventricle- ARV. The predicted segmentation labels were used to estimate cardiac physiological parameters and hand-crafted features. Random Forest-based feature importance analysis was performed to identify the most relevant features. The ensemble classifier system processed the features in two-stages for prediction of the cardiac disease as shown in the figure below.
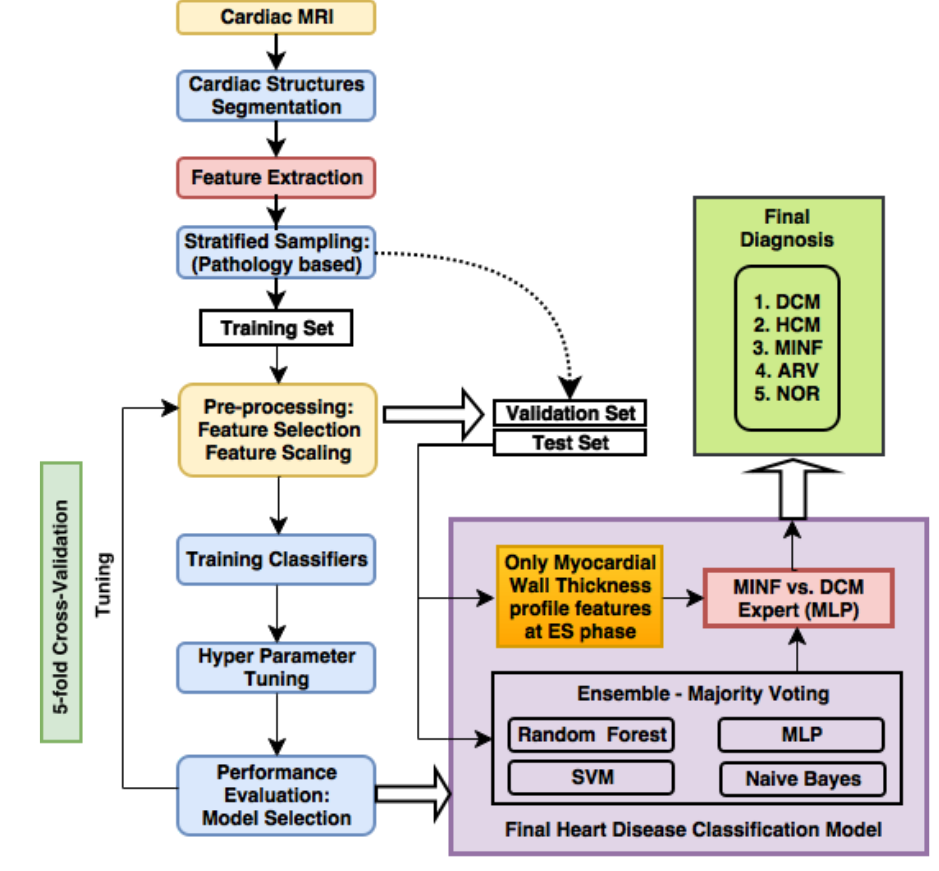 By following these steps in framework and network design, we achieved almost state-of-the-art performance on multiple cardiac segmentation datasets namely- (i) On STACOM ACDC-2017 challenge test set for segmentation task achieved a mean dice score of 0.94, 0.91 and 0.89 for the left ventricle, right ventricle and myocardium respectively and for automated cardiac disease diagnosis the accuracy was 100%, (ii) On STACOM LV-2011 test set the approach achieved 0.74 Jaccard index for myocardium segmentation. On the Kaggle challenge test set, the approach gave a continuous ranked probability score (CRPS) of 0.0127 for left ventricle volume estimation.
By following these steps in framework and network design, we achieved almost state-of-the-art performance on multiple cardiac segmentation datasets namely- (i) On STACOM ACDC-2017 challenge test set for segmentation task achieved a mean dice score of 0.94, 0.91 and 0.89 for the left ventricle, right ventricle and myocardium respectively and for automated cardiac disease diagnosis the accuracy was 100%, (ii) On STACOM LV-2011 test set the approach achieved 0.74 Jaccard index for myocardium segmentation. On the Kaggle challenge test set, the approach gave a continuous ranked probability score (CRPS) of 0.0127 for left ventricle volume estimation.
Use Case 2: Deep learning based framework for Whole-slide image segmentation
 Histopathology analysis is considered the gold standard in cancer diagnosis and prognosis. Whole slide imaging, i.e., the scanning and digitization of entire histology slides, are now being adopted across the world in pathology labs. Trained histopathologists can provide an accurate diagnosis of biopsy specimens based on whole slide images (WSI). Unfortunately, pathological analysis is an arduous process that is difficult, time-consuming, and requires in-depth knowledge. A study conducted by 2015 by Elmore group investigated the concordance of pathologists investigating biopsies of the breast. This study consisted of 115 pathologists across the United States and 240 biopsy specimens. It was found that pathologists disagreed with each other on a diagnosis 24.7% of the time on average. This high rate of misdiagnosis stresses the need to develop computer aided methods for histopathology analysis to aid the pathologists.
Histopathology analysis is considered the gold standard in cancer diagnosis and prognosis. Whole slide imaging, i.e., the scanning and digitization of entire histology slides, are now being adopted across the world in pathology labs. Trained histopathologists can provide an accurate diagnosis of biopsy specimens based on whole slide images (WSI). Unfortunately, pathological analysis is an arduous process that is difficult, time-consuming, and requires in-depth knowledge. A study conducted by 2015 by Elmore group investigated the concordance of pathologists investigating biopsies of the breast. This study consisted of 115 pathologists across the United States and 240 biopsy specimens. It was found that pathologists disagreed with each other on a diagnosis 24.7% of the time on average. This high rate of misdiagnosis stresses the need to develop computer aided methods for histopathology analysis to aid the pathologists.
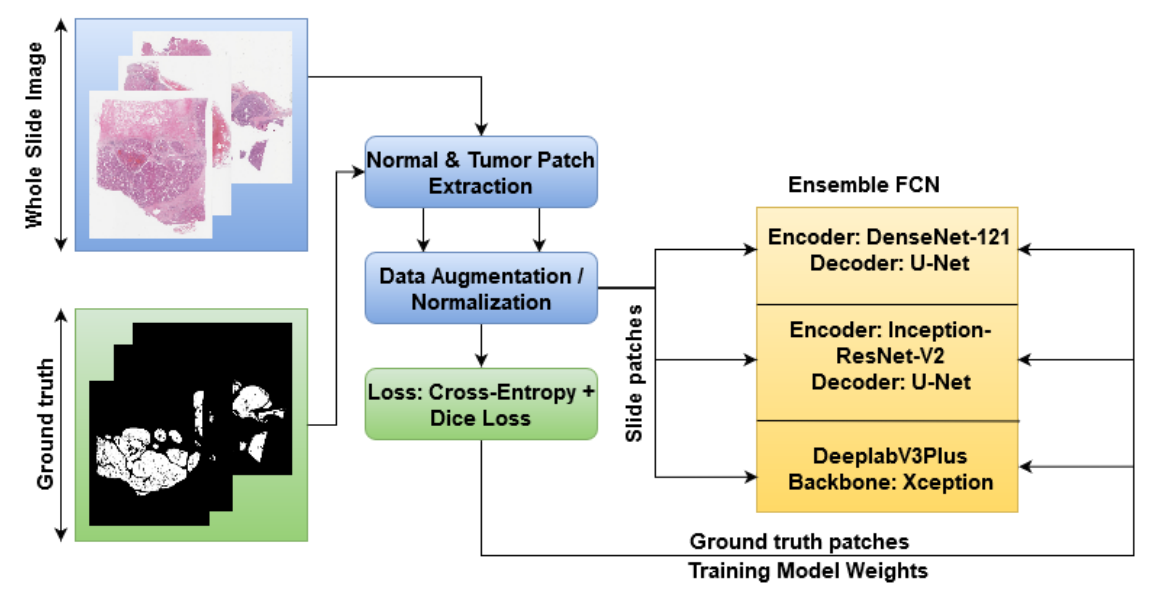
Though the computer aided solutions for histopathology comes with its own problems, the size and variability of images makes it difficult to train and analyse using CNNs. In the past decade deep learning methods have shown promising results in extracting and analysing tumorous tissue in WSI. But designing novel architecture and framework to do the same requires more effort and time. In one of our previous work we explored the CNN models for automatic segmentation and analysis of histopathology tissues. Here, we describe some best practices for developing the segmentation pipeline for large scale images. In our work we utilized multiple deep learning frameworks for segmentation (DenseUNet, InceptionUNet, and DeepLabv3) as described in the figure above. The task focused here is to differentiate tumor tissue from normal. Here are some useful steps which we followed:
- Ensembling based approach usually provides a high boost in performance. The above figure describes the same where the final segmentation is based on average probability maps obtained by individual models.
- In this experiment we used patches for training segmentation models, as histopathology images are generally (10k x 10k) which is hard to fit in GPU memory. For this patch based training the dataset is formed by randomly extracting patches from tissue regions from a histopathology images. Also the patches were equally sampled from tumorous and non tumorous regions to circumvent the problem of class imbalancing.
- Along with the proposed method of data sampling we even followed random augmentations including rotations, flips and color jitter, to make the model more robust to variations in a data.
- Now comes the inference stage, For efficient model inference we utilized some tricks to parallel infer multiple models for ensemble calculation over patches rather than an entire image. In inference setup the entire histopathology is first filtered with tissue masking step, where we identify the region in an image with tissue (normal and tumorous). After identifying tissue region, we sequentially sample patches with the overlap of 50% (this is done to eliminate sporous patch effects in the final image).
- Another step which we tried to enhance performance is to fine tune the model trained on CAMELYON data for other pathology tasks like digestpath and paip. Which resulted in the faster model convergence, it’s not ideal to use ImageNet weights in biomedical setup as most abstract features can not be reused.
- By following these steps in framework and network design, we achieved almost state-of-the-art performance on multiple histopathology datasets, including CAMELYON16, CAMELYON17, DigestPath, and PAIP. The scores achieved are listed as follows: (i) a FROC score of 0.86 for lesion detection on CAMELYON16 test data (n=139), a Cohen’s kappa score of 0.9090 for pN-staging on the CAMELYON17 test-data (n=500), (ii) a Jaccard Index of 0.75 for viable tumour segmentation and a metric score of 0.633 for the viable tumour burden estimation on PAIP test data (n=40) (second in the challenge), and (iii) a Dice score of 0.782 for tumour segmentation on DigestPath test data (n=212) (fourth in the challenge).
Conlcusion
In conclusion Deep Learning has a really nice application on medical images, In this blog we focused on segmentation problems, and provided some best practices in histopathology and cardiology MR images. Methods like patch based approach, multi-model ensemble, and fine tuning helps in boosting the performance of large scale image segmentation problems. That said, using pre-trained weights from ImageNet isn’t always beneficial.
Hope you find it useful, please get in touch if you find any difficulties…
Leave a Comment