Motivation
- Interpreting the decisions of AI models in the medical domain is especially important, where transparency and a clearer understanding of Artificial Intelligence are essential from a regulatory point of view and to make sure that medical professionals can trust the predictions of such algorithms.
- Understanding the organization and knowledge extraction process of deep learning models is thus important. Deep neural networks often work in higher dimensional abstract concepts. Reducing these to a domain that human experts can understand is necessary - if a model represents the underlying data distribution in a manner that human beings can comprehend and a logical hierarchy of steps is observed, this would provide some backing for its predictions and would aid in its acceptance by medical professionals.
- In this work the aim is to extract explanations for models which accurately segment brain tumors, so that some evidence can be provided regarding the process they take and how they organize themselves internally
Project Description
Level-1
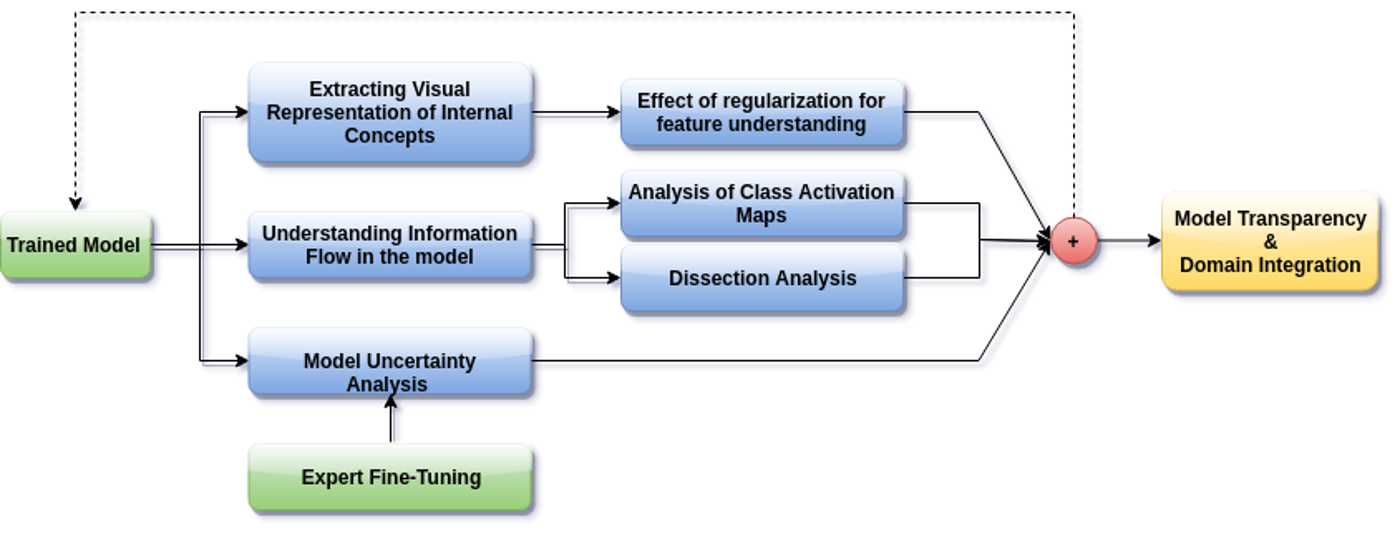
The accurate automatic segmentation of gliomas and its intra-tumoral structures is important not only for treatment planning but also for follow-up evaluations. Several methods based on 2D and 3D Deep Neural Networks (DNN) have been developed to segment brain tumors and to classify different categories of tumors from different MRI modalities. However, these networks are often black-box models and do not provide any evidence regarding the process they take to perform this task. Increasing transparency and interpretability of such deep learning techniques are necessary for the complete integration of such methods into medical practice. In this paper, we explore various techniques to explain the functional organization of brain tumor segmentation models and to extract visualizations of internal concepts to understand how these networks achieve highly accurate tumor segmentations. We use the BraTS 2018 dataset to train three different networks with standard architectures and outline similarities and differences in the process that these networks take to segment brain tumors. We show that brain tumor segmentation networks learn certain human-understandable disentangled concepts on a filter level. We also show that they take a top-down or hierarchical approach to localizing the different parts of the tumor. We then extract visualizations of some internal feature maps and also provide a measure of uncertainty with regards to the outputs of the models to give additional qualitative evidence about the predictions of these networks. We believe that the emergence of such human-understandable organization and concepts might aid in the acceptance and integration of such methods in medical diagnosis.
Level-2
The black-box nature of deep learning models prevents them from being completely trusted in domains like biomedicine. Most explainability techniques do not capture the concept-based reasoning that human beings follow. In this work, we attempt to understand the behavior of trained models that perform image processing tasks in the medical domain by building a graphical representation of the concepts they learn. Extracting such a graphical representation of the model’s behavior on an abstract, higher conceptual level would help us to unravel the steps taken by the model for predictions. We show the application of our proposed implementation on two biomedical problems - brain tumor segmentation and fundus image classification. We provide an alternative graphical representation of the model by formulating a concept level graph as discussed above, and find active inference trails in the model. We work with radiologists and ophthalmologists to understand the obtained inference trails from a medical perspective and show that medically relevant concept trails are obtained which highlight the hierarchy of the decision-making process followed by the model.
pip package
PyPi package for the following project can be found
here
-
Installation
pip install BioExp