Motivation
- Brain Tumors grow in various shape and texture, resulting in huge stochastic predictions by radiologists, automated tools using deep learning can help to reduce false positives in radiologists predictions
- In the hospitals with an inefficient workflow managements, tools like these will help radiologists to pick up critical cases
- Radiomic features from the tumor region provides lots of information about the tumor and helps in effective treatment
Project Description
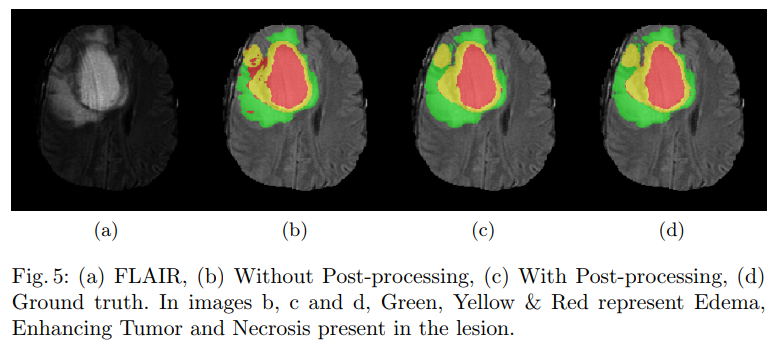
The accurate automatic segmentation of gliomas and its intra-tumoral structures is important not only for treatment planning but also for follow-up evaluations. Several methods based on 2D and 3D Deep Neural Networks (DNN) have been developed to segment brain tumors and to classify different categories of tumors from different MRI modalities. In this project, we utilize an ensemble of the fully convolutional neural networks (CNN) for segmentation of gliomas and its constituents from MRI. The ensemble comprises of 3 networks, two 3-D and one 2-D network. Of the 3 networks, 2 of them (one 2-D & one 3-D) utilize dense connectivity patterns while the other 3-D network makes use of the residual connection. Additionally, a 2-D fully convolutional semantic segmentation network was trained to distinguish between air, brain, and lesion in the slice and thereby localize the lesion the volume. Lesion localized by the above network was multiplied with the segmentation mask generated by the ensemble to reduce false positives. With the proposed technique we achieve the dice of 0.89 0.76, 0.76 on the whole tumor, tumor core and active tumor respectively. In this, we also provide the technique for uncertainty estimation, both model and data uncertainty which helps radiologists in making better decisions.
Open Source Packages
pip package
PyPi package for the following project can be found
here. The main motivation for PyPi python api to make it easier for other researchers to start with brain tumor analysis and develop this field.
Installation
pip install DeepBrainSeg
.deb package
Debian and .exe package for the project is under development. These applications are built considering radiologists as the end-user, here we try to assist radiologists to make quick and better decisions.
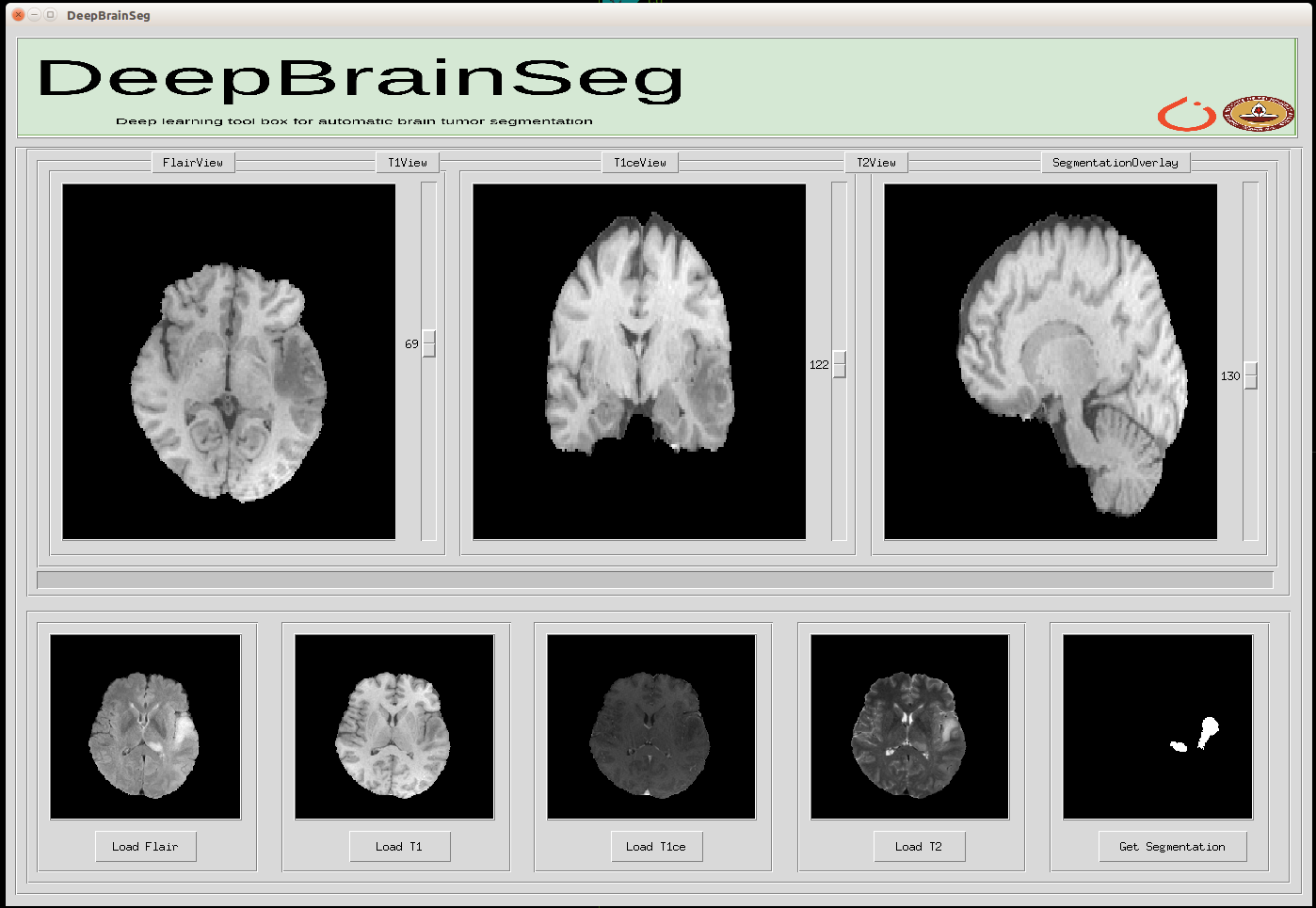
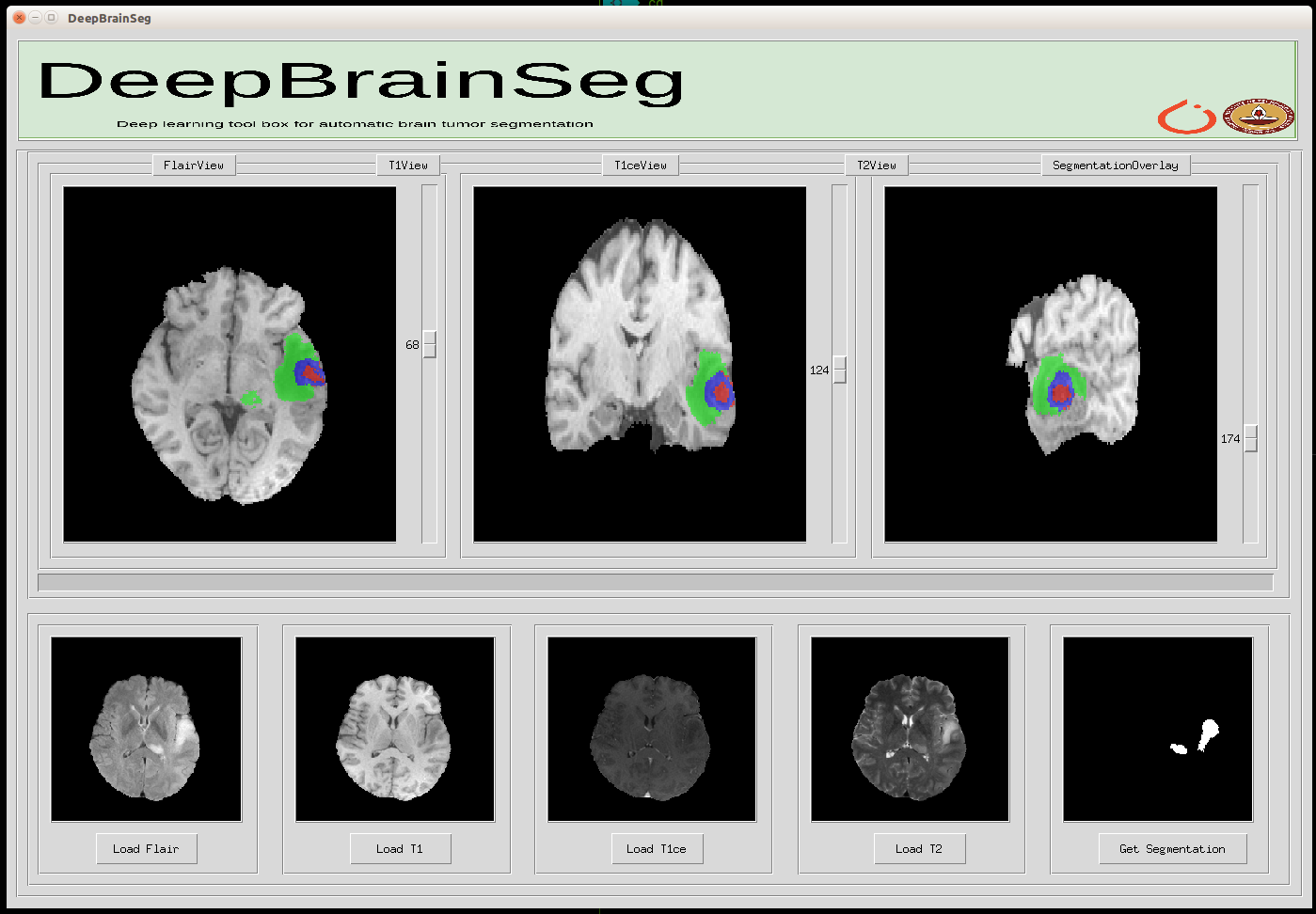
Some Images of our UI
-
Brain Images with four different MRI sequences
-
Brain Images with overlayed mask predicted by our algorithm
Github Link
Entire source code for python package and UI, is made open source so that other people can also contribute to the development of this project. Github repo can be found
here